Background
Synergy inference by data-driven network-based Bayesian analysis(SINBA) is used to bridge the novel discoveries from network-based systems biology to reverse pharmacology. SINBA will tremendously reduce the time and resource cost of conventional combination screening. SINBA also integrated a blood-brain barrier penetrated drug-target database, which will expedite the drug discovery for CNS tumors.
Installation
Require R >= 3.5.0.
- install from github
devtools::install_github("jyyulab/SINBA",auth_token = "your auth_token", lib="your lib path")- install from local file
pkg.dir <- "/Volumes/project_space/yu3grp/software_JY/yu3grp/git_repo/SINBA" #"/research_jude/rgs01_jude/groups/yu3grp/projects/software_JY/yu3grp/git_repo/SINBA"
devtools::install_local(sprintf("%s/SINBA_1.1.tar.gz",pkg.dir),lib="your lib path")Features
-
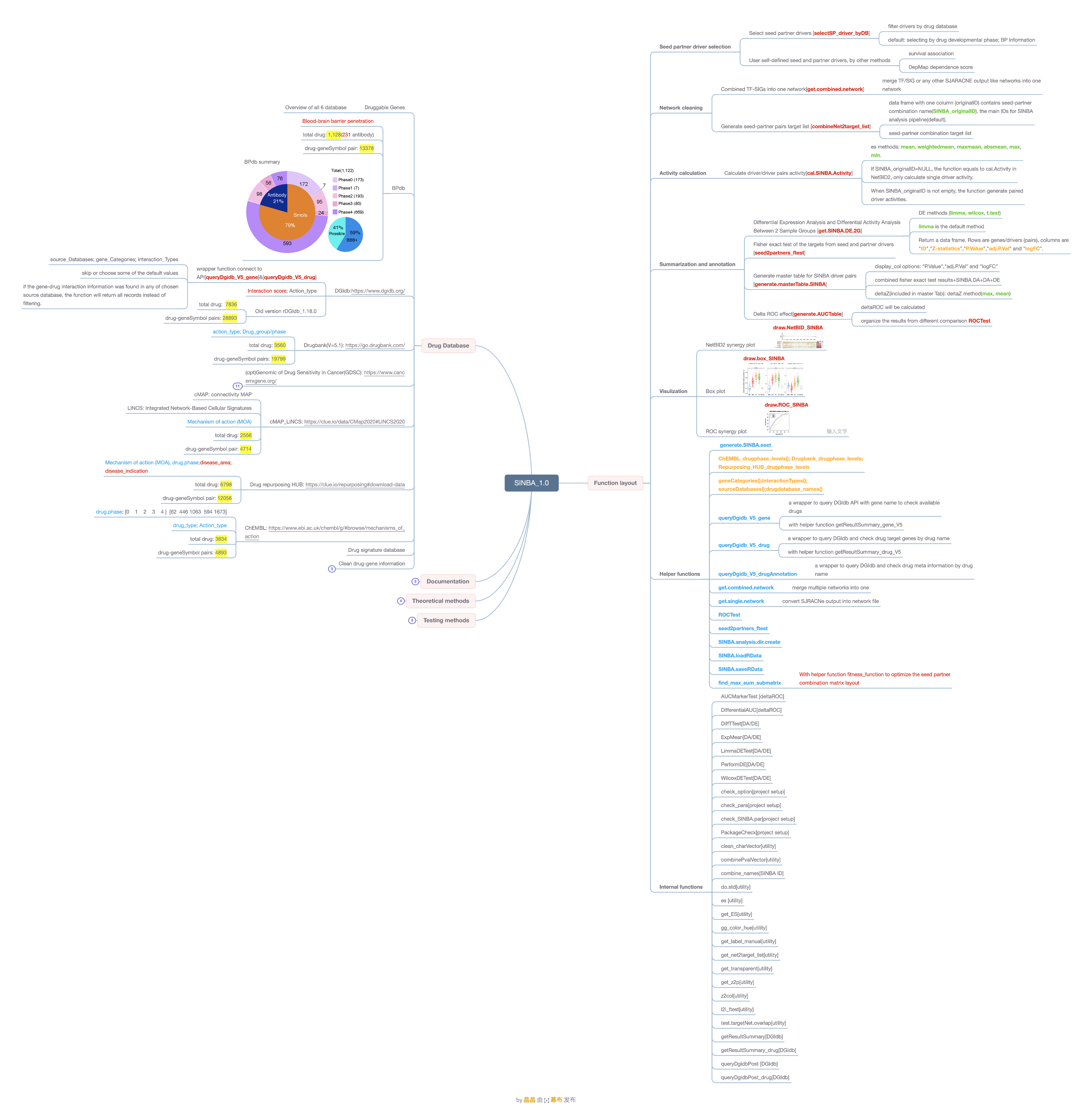
Comprehensive Drug-Gene Interaction Resources: Includes six curated drug-gene interaction databases, featuring the blood-brain barrier penetrant drug database (
BPdb), which supports the selection of CNS-specific drugs.
- Integrated Gene-Gene Interaction Network: Facilitates the inference of disease or subgroup-specific driver genes. These drivers may not have identifiable genetic alterations and can be “hidden” regulators affected by epigenetic, post-transcriptional, or post-translational mechanisms.
- Synergy Discovery Platform: Harnesses the strengths of in silico prediction and high-throughput screening to identify novel synergistic drug pairs.
References
| A. Khatamian, E. O. Paull, A. Califano and J. Yu Bioinformatics 2019: SJARACNe: a scalable software tool for gene network reverse engineering from big data. PMCID: PMC6581437 |
| X. Dong, L. Ding, A. Thrasher, X. Wang, J. Liu, Q. Pan, et al.Nat Commun 2023: NetBID2 provides comprehensive hidden driver analysis. PMCID: PMC10160099 |